Histone acetylation is a hallmark of chromatin that has an open structure that can be accessed by DNA and RNA polymerases as well as transcription factors, resulting in the activation of gene transcription (Filippakopoulos and Knapp, 2014). Correspondingly, histone methylation increases the basicity and hydrophobicity of histone tails and the affinity of certain proteins, such as transcription factors, toward DNA (Teperino et al., 2010), thus affecting the gene expression. In this database, we have collected 584 non-redundant protein data of 8 organisms including H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, A. thaliana, S. pombe and S. cerevisiae from the literature. The data are further classified into 15 families for histone acetylation writers, erasers and readers and 32 families for histone methylation writers, erasers and readers, respectively.
WERAM 1.0 is a comprehensive Eukaryotic Writers, Erasers and Readers protein of Histone Acetylation and Methylation system Database for 148 eukaryotic species. And here we provide two approaches for users to browse the database: (i) By species; (ii) By classifications.
| Browse by species | |||
| Animals | Plants | Fungi | |
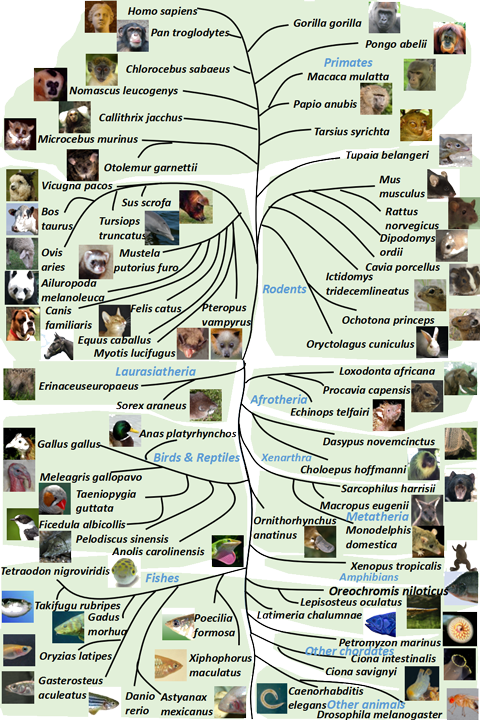 |
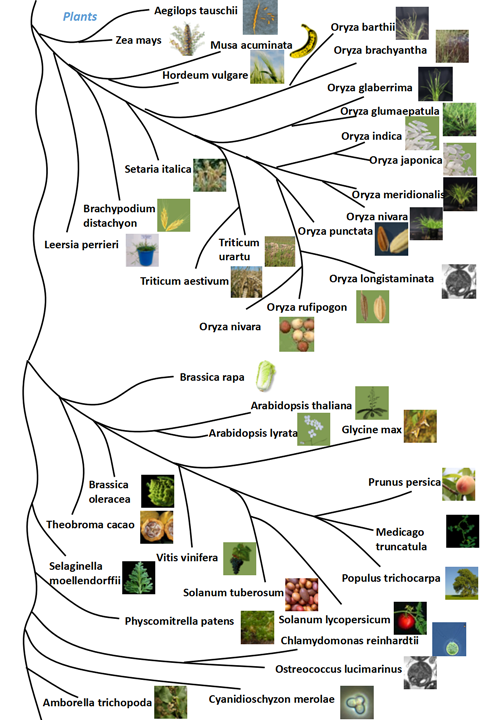 |
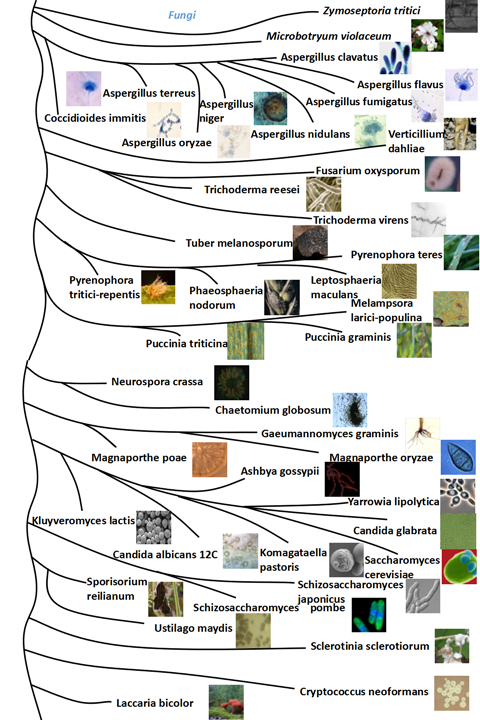 |
|
| Browse by classifications | ||||
| Histone Acetylation | Histone Methylation | |||
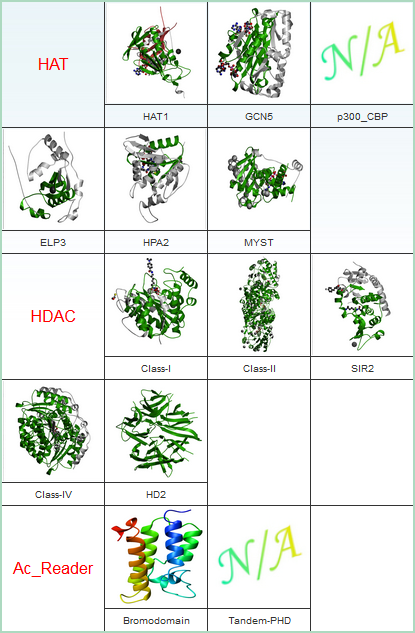 |
 |
|||
|
For publication of results please cite the following article:  WERAM: a database of writers, erasers and readers of acetylation and methylation in eukaryotes
WERAM: a database of writers, erasers and readers of acetylation and methylation in eukaryotesYang Xu, Shuang Zhang, Shaofeng Lin, Yaping Guo, Wankun Deng, Ying Zhang, and Yu Xue. Nucleic Acids Research. 2016, 45:D264–D270. [Abstract] [FREE Full Text] [PDF][Supplementary Data] |
|
|



